We are a team of computational and experimental biologists at St. Anna Children's Cancer Research Institute (CCRI). It is our goal to understand developmental processes in cancer biology by employing an integrative approach that combines stem cell biology with functional genomics.
We trace (aberrant) changes in cells during development.
Background: Hematopoiesis, immunity, and cancer
Cell Stem Cell 2016;
Science 2016;
Cancer Discov. 2019
We map regulatory networks that shape cell identity.
Background: Stem cell transcription factors
Cell Stem Cell 2012, 2013;
eLife 2017;
EMBO J. 2018
We develop tools that connect pieces of data into knowledge.
Background: Wet- and dry-lab tools and resources
Nat. Methods 2012;
Nat. Biotechnol. 2016;
Cell Stem Cell 2018
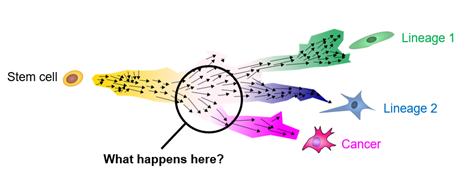
Our research currently focuses on understanding and modulating the earliest effects of cancer-linked mutations on development, tumorigenicity, and plasticity in embryonal tumors and other childhood cancers.
To this end, we use stem cell organoids and other experimental models and genomics technologies. Commun. Biol. 2025; Nat. Commun. 2024a; Nat. Commun. 2024b; bioRxiv 2025
Check out our latest pre-print: Montano, Mueller, et al.
Find out more about our research here! Do you have the know-how, creativity, and passion to join the quest? We'd like to hear from you!

