Taking apart pediatric cancer bit by bit
Goals
It is our goal to understand developmental processes in cancer biology. We track development of cancer cells in tumors, reconstruct these processes in experimental models, and characterize (epi)genomic changes that occur in time and space. With these data, we aim to decipher the syntax and semantics of development by formal abstraction into intelligible models. To this end, we combine developmental and cancer biology with high-throughput functional genomics assays (*-seq) and computational analysis. We like to collaborate and partner with research labs at CCRI and around the world to achieve these aims.
 +
+  +
+  = !
= !
Significance
Cancer is a disruption of orderly developmental processes – cancerous cells either fail to reach the destination of their developmental circuit or they revert after they had seemingly completed the track. By achieving a thorough understanding in individual cases and abstracting the findings in quantitative models, we help to elucidate new avenues for detecting (diagnosis), projecting (prognosis), and intercepting (therapy) with aberrant development in pediatric cancers.
Current funding:

ALSF (Crazy 8 Intiative)
We aim to find the origins of bone sarcomas. Info/press: ALSF, CCRI, ORF (in German).

St. Anna Kinderkrebsforschung
We investigate the development of embryonal tumors.
Examples of past projects:
Wishing for a cell
Stem cells can be made to differentiate into any kind of cell in the lab. Together with the Seruggia lab, we use chromatin approximation to find out how to make specific cell types well.
Read the pre-print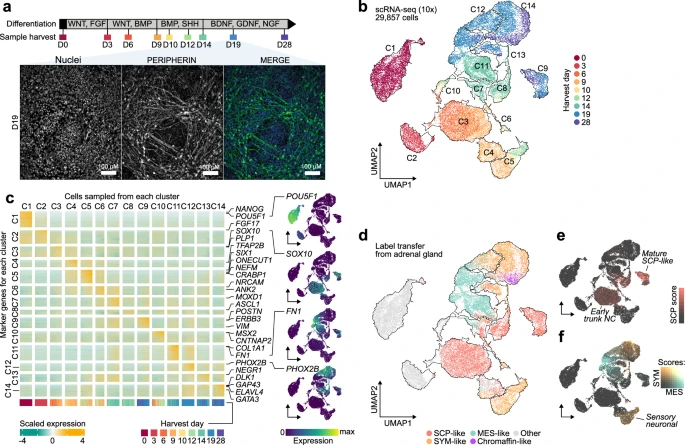
CNAs in neuroblastoma
Many pediatric cancers are associated with copy number alterations (CNAs). Making use of the Tsakiridis lab's stem cell model of the trunk neural crest, we investigated their role in neuroblastoma tumorigenesis.
Read the paper Get the code / data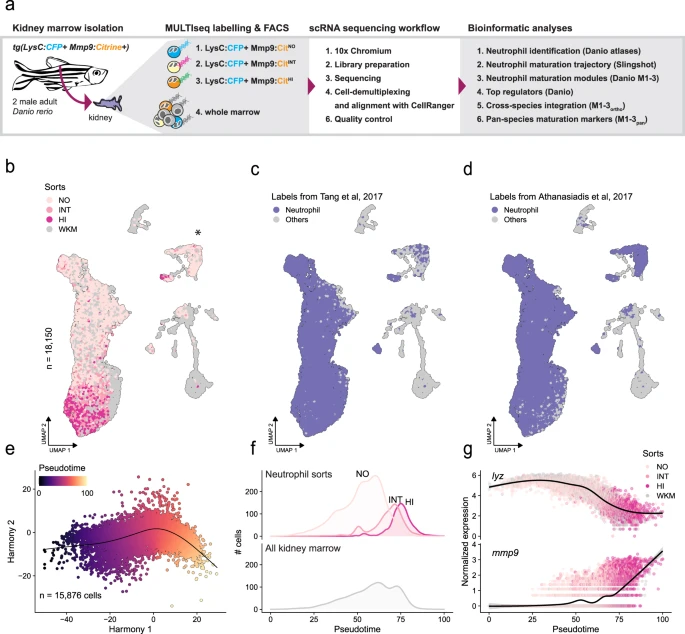
Neutrophil dev across species
Neutrophils are multifunctional immune cells involved in health and disease. Together with the Distel lab at CCRI we juxtaposed neutrophil maturation in zebrafish, mice, and humans.
Read the paper Get the code / data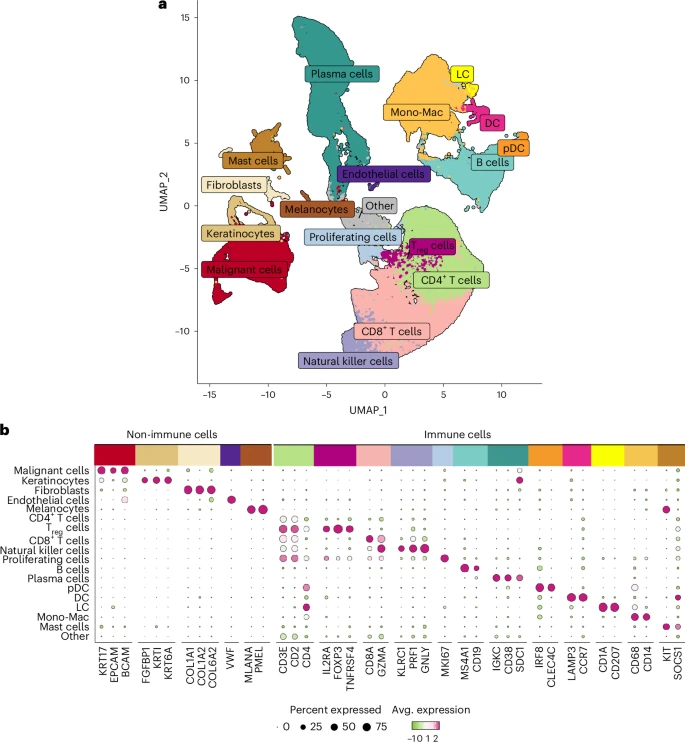
Oncolytic viruses in skin cancer
Basal cell carcinoma is the most common cancer worlwide. We participated in the analysis of data from a clinical trial organized by the Hoeller lab at the Medical University of Vienna.
Read the paper Get the code / data
SARS-CoV-2 mutations
Our small contribution to the pandemic: We teamed up with researchers at CeMM and the Medical University of Vienna to investigate the effect of SARS-CoV-2 mutations on CD8+ T cell immunity.
Read the paper Get the code / data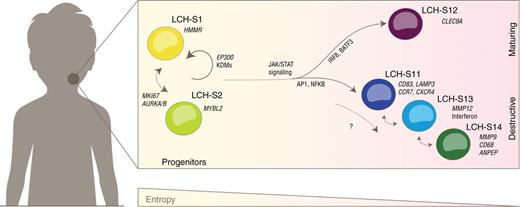
Intra-tumor plasticity in LCH
Langerhans cell histiocytosis is an enigmatic neoplasm with highly variable clinical presentation. Together with CeMM, we charted LCH at single-cell resolution and unraveled an unexpected hierarchy.
Read the paper Get the code / data
Finding enhancers
Enhancers are genetic elements that regulate gene expression. Our study in collaboration with Edinburgh and Rotterdam used a genome-wide assay to explore enhancer activity in pluripotent stem cells.
Read the paper Get the code / data
Kinases & blood
Cells are shaped by the enzymatic activity of kinases. Two recent contributions implicate MAPK in blood stem cells, and JAK+Aurora in mutant immune cells -- with impacts on the transcriptome and epigenome.
MEKi & HSCs STAT5 Neoplasia
Epigenetics of hematopoiesis
The hierarchical development of the human blood system is a fascinating process. In the Blueprint Consortium, we mapped epigenetic (DNA methylation) changes during hematopoiesis globally.
Read the paper Get the data
Macrophage development
Tissue-resident macrophages are key players in immunity, development, and homeostasis. We teamed up with the Geissmann and Schultze labs to trace the embryonic orgins of these cells.
Read the paper Get the data
Biomarker benchmark
DNA methylation is a stable epigenetic mark well suited for biomarker assays. We conducted a large benchmark with 16 labs worldwide to compare platforms for clinical translation.
Read the paper Get the code
Regulation of pluripotency
We have investigated multiple core regulators of mammalian pluripotency to discover what makes ESCs tick.
Esrrb Oct4/Nanog Sox2/Oct Sox2/Nanog Sox Esrrb-loss
Streamlined NGS analysis
To ease access and reuse of functional genomics data, we developed a web-based software suite and database that streamlines NGS data analysis.
Read the paper Read the other paper

